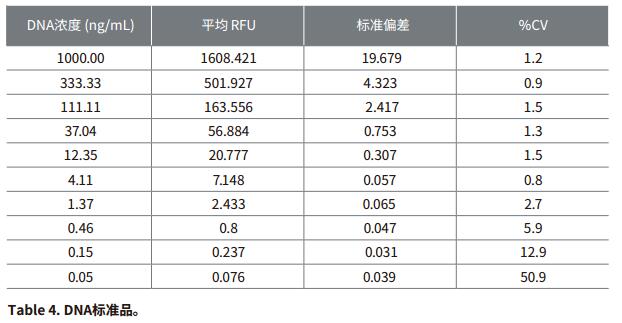
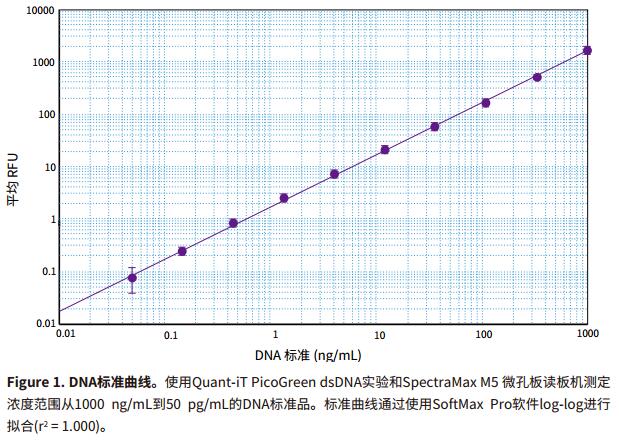
Introduction Double-stranded DNA is typically quantified by measuring the 260 nm absorbance of the DNA solution using a microplate reader. However, this method typically only detects a concentration of at least 250 ng/mL on a microplate reader. For biological applications involving small volume samples, such as next-generation sequencing and amplification product quantification, a more sensitive approach can be met. Life Technologies' Quant-iT PicoGreen dsDNA assay is more specific for DNA and 1000 times more sensitive than traditional light absorbing methods. The product information is nominally performed on a microplate using a single dye concentration with a dynamic range of 250 pg/mL to 1000 ng/mL. 1 This application note demonstrates the ability to stably detect double-stranded DNA as low as 100 pg/mL using the Molecular Devices SpectraMax® Fluorescence Microplate Reader and the Quant-iT PicoGreen assay. In order to maximize the sensitivity of the experiment, it is necessary to use optimal excitation and emission wavelengths. Unlike color filter-based plate readers, the SpectraMax Microplate Reader's dual grating allows the user to select any wavelength within the instrument's nominal range. It is important to determine the excitation and emission wavelengths that produce the best experimental dynamic range, as this will be somewhat different from filter-based readers. The optimal excitation and emission wavelengths of the SpectraMax M5 Fluorescence Microplate Reader are as follows: 490 nm excitation and 525 nm emission, 515 nm emission intercept filter (note that these wavelengths may be related to color filter based reading in Quant-iT PicoGreen product brochures) The wavelength of the machine is different). There are preset programs in the SoftMax® Pro software to capture, display and analyze data from the SpectraMax series of readers. Advantage - Sensitive fluorescence quantification of DNA as low as 63 pg/mL material - Quant-iT PicoGreen dsDNA kit containing l ambda DNA standard (Life Technologies Cat. #P7589 or P11496) method Instrument settings - Turn on the microplate reader. Prepare the experiment The experimental method follows the guidance in the product information page of the QuantiT PicoGreen dsDNA reagent and kit, except that the experimental volume is reduced from 2.0 mL to 200 μL in proportion to the 96-well microplate format. - 1XTE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was prepared by using a concentration buffer in a 20-fold dilution kit without DNase water. Note: In some cases it may be necessary to make a mark using a DNA type similar to the DNA to be tested. - High concentration range of standards can be prepared from 1 ng/mL to 1 μg/mL, and low concentration range can be prepared from 25 pg/mL to 25 ng/mL. For high concentration range calibration, dilute according to the dilution scheme in Table 2; for low concentration range calibration, 40-fold dilution of 2 μg/mL solution to prepare 50 ng/mL solution, refer to the dilution scheme in Table 3. Note: This application note uses a series of calibrations from 1 μg/mL to 50 pg/mL. Reading microplate - Make sure the purple board adapter (if required by your reader) is placed in the plate holder of the microplate reader. analyze data - After the microplate is read, the relative fluorescence units (RFUs) are displayed in the plate block. The data in the group table set by the template is analyzed. For a set of data from a representative set of tables, see the Table 4 Results section. result DNA standards at concentrations ranging from 1 μg/mL to 50 pg/mL were determined using the Quant-iT PicoGreen dsDNA assay and the SpectraMax® M5 Microplate Reader (Molecular Devices' other fluorescent microplate readers gave similar results). The SoftMax® Pro software repeats the automated calibration of each set of standards and calculates the average RFU, standard deviation, and %CV. Fitting the standard curve using the SoftMax Pro software loglog curve fit (see Figure 1.) The sensitivity observed in the 96-well plate microplate format was as low as 63 pg/mL and the standard detection limit was calibrated by the plate standard deviation of three times the standard deviation. . This value is significantly lower than the nominal 250 pg/mL in the QuantiT PicoGreen experimental product specification. Figure 2 shows the low concentration end standard curve. The linearity of the entire range of the calibration is very good. in conclusion The Quant-iT PicoGreen dsDNA assay is a fast and sensitive assay for double-stranded DNA when tested on a SpectraMax microplate reader using SoftMax Pro software. The analytical capabilities of SoftMax Pro software provide quantitative analysis in a convenient, customizable report format. Programs preset in the software make quick experimental setup possible. Additional solutions from Molecular Devices SoftMaxProGxP software provides additional tools and features for GLP/GMP customers who must demonstrate compliance with FDA21CFR Part11. Optimized hardware and software verification tools to accelerate and simplify the verification process, making it easier to demonstrate compliance with data collection and analysis. For higher throughput requirements, Molecular Devices' 'StakMax® Multiwell Plate Operating System integrates the SpectraMax reader and automates the processing of 20, 40, or 50 batches of microplates. For customers who need to detect extremely low levels of DNA, Molecular Devices offers the Threshold® System and Threshold Total DNA Test Kit. The Threshold system detects and quantifies picogram-level contaminants, including total DNA host cell proteins, bovine source contaminants (such as BSA, IgG, insulin, transferrin), proteins A and G, and any antibody that binds to antibodies. Unique protein. For more information, please visit reference 1. Molecular Probes Data Sheet MP 07581 (20 December 2005): Quant-iT PicoGreen dsDNA Reagent and Kits. Medical Surgical Masks Disposable medical masks are three-layered regular authentic children and individually packaged for adults. Only if there is no peculiar smell, it is a real mask. Upgraded, widened and thick ear straps, after authoritative testing, can bear weight, high elasticity, no ears, no break, and comfortable to wear. High-elastic ear straps, comfortable and non-straining ears, the surface is evenly densely covered with pores, comfortable and breathable, low sensitivity and no irritation. There is no burr, and it is our responsibility to check health. China Protective Mask,Disposable Mask supplier & manufacturer, offer low price, high quality Disposable Medical Dust Mask,Medical Disposable Face Mask, etc. Protective Mask,Disposable Mask,Disposable Medical Dust Mask,Medical Disposable Face Mask KUTA TECHNOLOGY INDUSTRY CO.,LIMITED , https://www.kutasureblue.com
- Linear dynamic range spans four orders of magnitude
- Simple analysis of results through preset programs in SoftMax Pro software
- Brown 96-well plate (Greiner Bio-One, Cat#655096)
- brown or amber (light-proof tube)
- Molecular Devices Microplate Reader with Fluorescence Detection Mode:
- S pectraMax® M3/M4/M5/M5e(cat. #M3 or #M4 or #M5 or M5E)
- SpectraMax® i3x (cat. #i3x)
- S pectraMax® M2/M2e(cat. #M2 or M2E)
- Gemini XPS/EM (cat. #XPS or EM)
- FlexStation® 3 (cat. #FLEX3)
- FilterMax F3/F5 (cat. #F3 or F5)
- S pectraMax® Paradigm(cat. #PARADIGM)
- SoftMax® Pro Software (Molecular Devices)
- Launch the SoftMax Pro software and open the PicoGreen fluorescence program in the Protocols drop-down menu. The optimal settings for the experiment are included in the program (see Table 1). Select the hole you want to read and the type of the experiment board by clicking on "Settings" and on the left side of the screen.
- Click on the "Template" button to open the window where you can assign the holes of the perforated plate to the corresponding groups of the preset template. You can use the drop-down menu to select the appropriate template group. There are several preset template groups in the fluorescence program, including Standards, Unknowns, and Unknowns_NoDiln (corresponding to diluted samples). Assigning holes to the group of the current template groups the data obtained when the corresponding microplates in the program group table are read.
- An aqueous phase working solution was prepared using a 200-fold dilution of concentrated Quant-iT PicoGreen reagent DMSO solution in TE buffer (prepared above). It is recommended to prepare this solution in a plastic container instead of in a glass container because the reagent may be adsorbed on the glass surface. Protect the reagents from darkness by using amber or brown tubes or wrapping them in metal foil. It should be used within a few hours after preparation of the reagents.
- DNA standard curve: A 2 μg/mL dsDNA stock solution was prepared using TE buffer. The ambda DNA standard provided in the kit can be made into a 2 μg/mL solution by diluting 50 times with TE. 


- Transfer 100 μL of each well to a black 96-well microplate using a pipette, preferably 3 replicates. Make sure to add TE (no DNA) to the well as a blank well.
- Add 100 μL per well of Quant-iT PicoGreen Reagent aqueous working solution. Mix by vortexing or shaker at room temperature for 2 to 5 minutes.
- Click the Read button in the SoftMax Pro software. The instrument reads the board and the relative fluorescence units are displayed in the board section of the program.
- Assigning the specified standard to the template will automatically fit the curve to the curve (and display it in the standard group table) in the program. The default setting is a linear curve fit, but a loglog curve fit can be used when fitting a wide range of markers. You can select a curve fit in the Curve Area Curve Fit drop-down menu.